GenomeView 2250 changelog
Fri, 08/09/2013 - 20:04 — Thomas AbeelThere have been a number of changes since the last update that was posted.
First of all, we've switched our versioning system to git, this should make it more straight-forward for more contributors to join.
We've added some extra instructions to be used in session files, this should make session files more versatile.
Redesign rendering of read tracks which should speed up rendering significantly.
Added wild cards to motif searches.
Added code automatic plugin installation option in session files.
GenomeView on the cover of new book edited by Stuart Brown
Fri, 02/15/2013 - 23:18 — Thomas Abeel Book cover
Book cover
GenomeView is featured prominently on the cover on the new book edited by Stuart Brown, Informatics for next generation sequencing, published by CSHL press.
GenomeView 2020 changelog
Wed, 11/21/2012 - 21:54 — Thomas Abeel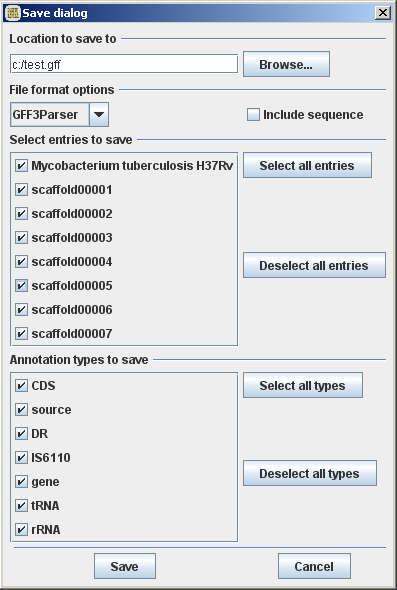
Changes in GenomeView
- revamped save dialog to be more comprehensive and intuitive
- ghost outline for total coverage is lighter
- comparative annotations show with intron-exon structure
- use synonym service to find a common name for data entries
- remember last loaded data an location for next start-up
- confirm dialog when closing GV
- location and individual config options can be specified in sessions files
Changes in JAnnot
- fixed issue in EMBL parser for valueless keys
GenomeView 1983: strand-specific coverage plots
Fri, 10/26/2012 - 21:32 — Thomas AbeelBiggest novelty over the past few months is the ability to create and visualize strand-specific coverage plots.
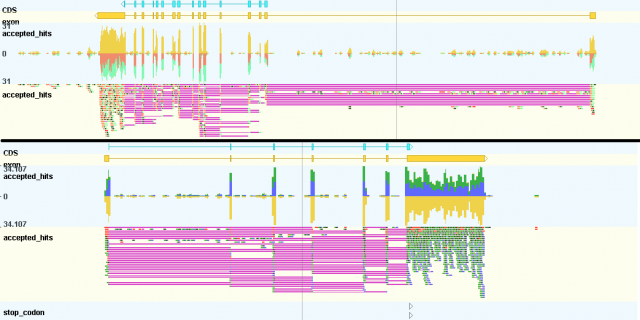
To create these files, you can use the latest version of tdformat
I've started a twitter account to make smaller announcements as they come online, so feel free to follow @GenomeView to stay up to date on any new developments.
GenomeView first paper published
Mon, 11/21/2011 - 17:37 — Thomas AbeelWe are pleased and proud to announce that the first GenomeView manuscript has (finally) been published.
GenomeView: a next-generation genome browser
Thomas Abeel; Thomas Van Parys; Yvan Saeys; James Galagan; Yves Van de Peer
Nucleic Acids Research 2011; doi: 10.1093/nar/gkr995
[PubMed] - [Nucleic Acids Research]
GenomeView 1805: improved multiple alignments and support for bigWig
Tue, 10/11/2011 - 19:59 — Thomas AbeelThe recent releases between 1682 and 1805 have added a number of new feature to GenomeView, a number of improvements to existing features as well as a whole set of bug fixes.
From now on, the bigWig format, that's been in use over at UCS, is supported by GenomeView. bigWig is used to store large continuous value data files.
The save and export functionality has been expanded to allow more fine grained control of what data has to be saved.
GenomeView wins 'Most creative visualization' award in the Illumina iDEA challenge
Fri, 07/08/2011 - 23:23 — Thomas Abeel Most creative visualization award @ Illumina iDEA challenge
Most creative visualization award @ Illumina iDEA challenge
Illumina's technologies have the potential to help us understand diseases and the living world around us at the molecular level. Unlocking that knowledge will enable radical improvements in human health and quality of life over the next decade. A key component to achieving this vision lies in empowering and accelerating the analysis, visualization and interpretation of the data being generated with these technologies. With this in mind, Illumina is announcing the iDEA (Illumina’s Data Excellence Award) Challenge.
GenomeView won the Most Creative Visualization award.
GenomeView 1682: integrated indexing, user interaction and reduced memory consumption
Fri, 07/08/2011 - 02:00 — Thomas AbeelRelease 1682 is again a step forward in making GenomeView as user-friendly as possible.
Major new features:
- Automagic index creation: If you try loading a file that is not yet indexed, GenomeView will now ask to do it for you.
- Speed and memory improvements: We have significantly optimized the way annotation feature and sequences are handled so it is easier to load monstrous draft genomes with 65,000 contigs and 500,000 annotation items without needing a gigantic computer.
GenomeView 1600: bugs, convenience and performance
Thu, 06/09/2011 - 15:14 — Thomas AbeelBased on user feedback we have been able to fix another batch of bugs as well as introduce a number of convenient new features. Keep the reports coming, we appreciate all feedback.
Selected changes
GenomeView now remembers in which order tracks are, and will no longer jumble them if you switch chromosomes or when you restart GenomeView. It also remembers which track you made invisible.
The pileup and formats are now also loaded using background threads, which drastically improves the responsiveness of the GUI.
GenomeView 1578: maintenance and usability
Fri, 05/20/2011 - 22:55 — Thomas AbeelBased on user feedback a number of bugs have been fixed. Furthermore, we have tried to improve usability with a number of small improvements.
Release 4 of the genome of Rattus norvegicus is now included in the Genome Explorer.


