GenomeView 1983: strand-specific coverage plots
Biggest novelty over the past few months is the ability to create and visualize strand-specific coverage plots.
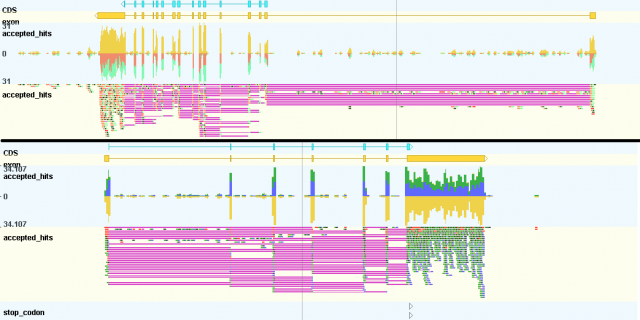
To create these files, you can use the latest version of tdformat
I've started a twitter account to make smaller announcements as they come online, so feel free to follow @GenomeView to stay up to date on any new developments.
There have been quite a few smaller improvements between this version and the previous announcement of 1805, which includes quite a few improvements that already made it into the first paper:
- GTF is now supported as a full format with its own parser
- fixed several graphical glitches in the gene structure view
- added a simplified color scheme for NGS reads
- visual indication of reads that have their mate mapping to another chromosome
- added all bacterial genomes from NCBI as a repository
- fixed graphical glitch when reads have soft-clipping at the beginning
- More checks on newly created features so they are not out of bounds of the reference
- User can no longer selection sequence outside the reference range
- Improved display labels on tracks
- Track aliases
- fixed graphical glitches in coverage plots
- File URLs that have an '=' sign get properly processed
- log-scaling for coverage tracks
- Improved detection of species/contig distinction in multiple alignment to reduce the number of entries.
- support multi-column wig files
- loading files from a session is more suble, no longer a bright green pop-up in the middle of your screen that blocks everything else.
- parser detection also works for sessions files, they can now be loaded as regular files
- tracks now have individual configuration menus
- have to option to automatically normalize by the mean coverage for coverage plots
- fixed one-off in search results on the reverse strand
Technical improvements:
- more and better logging so it's easier to help you when things break
- stream caching so the server is only contacted once per file
- better propagation of javascript instructions when multiple GenomeView instances are running
- made javascript instructions more robust and detect duplicate and garbled instructions
- fixed some memory leaks of objects that were not released when data unloads



